by
John R. Fischer, Senior Reporter | April 26, 2019
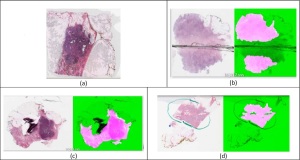
A new quality-control solution validates
if slides are of accurate or poor
quality for making diagnoses
With no standards laid out for how to sort and digitize tissue slides, the mixing of accurate and poor quality samples can create problems in the diagnosis process. For instance, such an arrangement could potentially confuse or mislead a computer program that is being trained to identify what a cancerous cell looks like.
Bioengineering researchers at Case Western Reserve University are setting out to change this with the development of a quality control program for validating the quality of digital images used in diagnostic and research settings. The name of their project is "HistoQC," for "histology," and "Quality Control."
"Slide digitization is still relatively new. As digital pathology becomes more prevalent and gains more clinical traction, the problem of standards for quality control and assurance will become more critical," Anant Madabhushi, the F. Alex Nason Professor II of biomedical engineering at the Case School of Engineering, told HCB News. "HistoQC attempts to provide a tool to help address this impending need for the pathology community."



Ad Statistics
Times Displayed: 2095
Times Visited: 26 Keep biomedical devices ready to go, so care teams can be ready to care for patients. GE HealthCare’s ReadySee™ helps overcome frustrations due to lack of network and device visibility, manual troubleshooting, and downtime.
Imperfections in slides can be caused by a variety of factors, from air bubbles, smears and ragged cuts, known as knife chatter, during slide preparation, to blurriness or brightness issues that take place during digitalization.
The open source application was developed by Andrew Janowczyk, a senior research fellow in Madabhushi's Center for Computational Imaging and Personal Diagnostics and a bioinformatician at the Swiss Institute of Bioinformatics, who was surprised by the number of poor-quality slides within the Cancer Genome Atlas, which holds more than 30,000 tissue slides of cancer samples. Of the 800 he reviewed, about 10 percent had problems.
Supported through a three-year, $1.2 million grant from the National Cancer Institute, HistoQC relies on a series of measurements and classifiers to flag and alert users to corrupted images, while helping to identify ones that will aid in diagnoses.
These specific classifications include a series of different types of artifacts, such as breaks in glass, hair, follicles, tissue folds, and blurriness in images, and is expected to expand, as the quality-control tool is designed for newer images and will eventually be trained to identify new classifications on those.
"Most critically, HistoQC can help identify based on image statistics of a slide, whether it is potentially an 'outlier' or an 'exemplar' of an odd-looking slide. This information from the user interface can be used to flag those slide images that are potentially 'off' and need to be either re-scanned or recreated," said Madabhushi. "This is important since it is critical for these "artifact laden" slide images to be identified prior to going to the pathologist for diagnostic interpretation. Rather than having the pathologist identifying the slide images as problematic and then having to send them back, the histotech can catch the faulty slide images early and reduce the number of poor quality slide images that land on the pathologist's desk."
Aiding the project are research partners from University Hospitals, the Perelman School of Medicine at the University of Pennsylvania and the Louis Stokes Cleveland VA Medical Center.
The findings were published in the
Journal of Clinical Oncology Clinical Informatics.

